Evolution and adaptation of microbial genomes
We apply experimental, comparative and computational approaches to examine the evolution and adaptation of microbial genomes.
Due to their complex and varied interactions as pathogens and commensal constituents of mammalian hosts, our work focuses on genome evolution within enteric bacteria, including E. coli, Shigella and Salmonella. These studies fall into four general areas.
Click on images for further information.
Dynamics of bacterial genomes
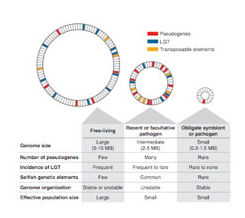
Numerous opposing forces contribute to large-scale differences the size, contents and organization of the bacterial genomes. Such alterations, whose characterization was limited previously to the few human pathogens amenable to genetic manipulation, have become increasingly evident with the recent availability of whole genome mapping techniques and of complete genomic sequences. Our research investigates the accumulated differences promoted by gene transfer and gene loss, and how the genome restructuring caused by these processes affect bacterial lifestyle.
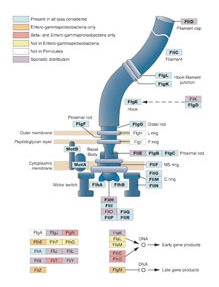
Origins and roles of novel bacterial traits
Within most bacterial genomes, the majority of genes is of unknown function and these genes are likely responsible for vast diversity in bacterial lifestyles and metabolic functions observed in the microbial world. Using a combination of experimental and bioinformatic methods, our research addresses two questions:
- What are the origins and roles of sequences that have been acquired through horizontal transfer?
- What is the function of genes for which no role can be assigned by conventional homology-based and genetic approaches?
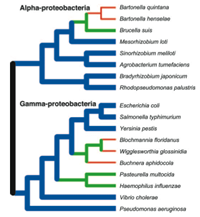
Phylogenetic classification and the history of bacterial genomes
The use of universally distributed molecular characters to resolve the relationships among organisms has been particularly informative in micro-organisms, in which the lack of morphological characteristics and lack of a robust fossil record have hinder previous attempts to determine their evolutionary history. In addition, recent studies provide clear evidence that lateral gene transfer is common among bacteria with the result that different portions of the genome have very different histories.
Our research in this area generates and examines nucleotide sequences for a large set of genes that are universally distributed among Bacteria to determine:
- how well the phylogenetic trees based on small different genes represent the relationships of organisms;
- whether organisms contain a core set of genes that are never subject to transfer;
- which genes are transferred among lineages, and what are the phylogenetic limits to lateral gene transfer;
- whether the genes subject to lateral transfer functionally related or genetically linked; and
- what factors determine the amount of acquired DNA in a genome.
Genomic analysis of diversity within bacterial communities
The tissues and cells of eukaryotic hosts serve as the environment for vast numbers of bacterial species, with associations ranging from obligatory and beneficial to antagonistic and parasitic. We are exploring the assemblages of bacteria inhabiting the human GI tract to determine what properties determine the species composition, genetic characteristics and persistence and of bacteria within the intestinal flora. In a related project, we are developing methods to isolate individual constituents of bacterial assemblages for genomic characterization.