Nancy A. Moran
Research
CURRENT RESEARCH
PAST RESEARCH
- Elucidating the molecular bases of species interactions in host-associated bacterial communities
- Role of Gut Microbiota in Honey Bee Health
- Coevolutionary dynamics in an obligate insect symbiosis
- Colonization by a Co-evolved Gut Community
- Dual Obligate Intracellular Symbionts
- Control of Bee Behavior by Stably Engineered Gut Microbial Communities
- Dimensions of Biodiversity: the Gut Microbiota of Bees
- Environmental Genomics of Symbionts in Pea Aphids
- Genomics of Bacterial Symbionts of Plant Sap-Feeding Insects
- Biocomplexity in the Environment
- Bacterial Endosymbiont Diversity in Drosophilla
- Biocomplexity of Symbiotic Bacteria
- Genomic Evolution of Buchnera
- Evolutionary Dynamics of Endosymbiont-Borne Adaption on Aphids
- Molecular Phylogenetics of Sternorrhyncha
- Phylogenetics of Aphids
- Genetically Variable Complex Life Cycles in Heterogeneous Environments
Research Projects in the moran lab
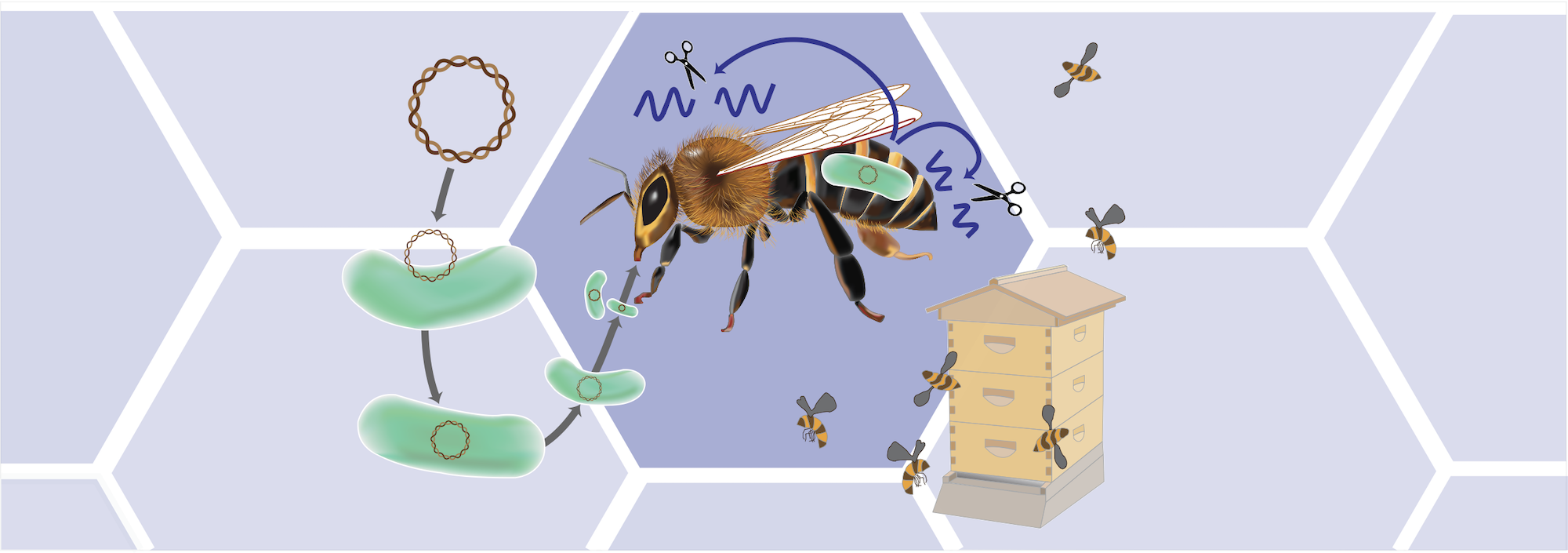
Bee Functional Genomics Using Engineered Symbionts
(PI is Jeffrey Barrick) (Co-Principal Investigator is Nancy Moran)
 Insects are among the most widespread and diverse animals on our planet. They have critical roles in natural ecosystems and agriculture and have evolved unique biomaterials and lifestyles. Scientific tools for studying the functions of the genes responsible for these traits are well-established for only a few types of insects, such as fruit flies. Genetic tools do not exist for millions of other species. Honey bees and bumblebees are economically important as widespread pollinators of food crops and are scientifically interesting due to the complex social behaviors observed in bee colonies. Currently, there are few effective tools for studying the functions of bee genes. This project will develop and disseminate a toolkit that allows researchers to alter the expression of bee genes by engineering their native symbiotic gut bacteria. This technology will enable studies of how specific genes contribute to bee physiology, development, and behavior. This work will contribute to understanding bee ecology and health in ways that are expected to benefit biodiversity and US food security in the long term. The technology for engineering symbiotic bacteria is expected to be widely applicable to studying other insect species. These research and outreach activities will be integrated with education by supporting two experiential learning courses that are part of the Freshman Research Initiative program at The University of Texas at Austin. Supporting this program will foster the development of a diverse science and technology workforce by involving underrepresented and first-generation college students in genuine research experiences.
Insects are among the most widespread and diverse animals on our planet. They have critical roles in natural ecosystems and agriculture and have evolved unique biomaterials and lifestyles. Scientific tools for studying the functions of the genes responsible for these traits are well-established for only a few types of insects, such as fruit flies. Genetic tools do not exist for millions of other species. Honey bees and bumblebees are economically important as widespread pollinators of food crops and are scientifically interesting due to the complex social behaviors observed in bee colonies. Currently, there are few effective tools for studying the functions of bee genes. This project will develop and disseminate a toolkit that allows researchers to alter the expression of bee genes by engineering their native symbiotic gut bacteria. This technology will enable studies of how specific genes contribute to bee physiology, development, and behavior. This work will contribute to understanding bee ecology and health in ways that are expected to benefit biodiversity and US food security in the long term. The technology for engineering symbiotic bacteria is expected to be widely applicable to studying other insect species. These research and outreach activities will be integrated with education by supporting two experiential learning courses that are part of the Freshman Research Initiative program at The University of Texas at Austin. Supporting this program will foster the development of a diverse science and technology workforce by involving underrepresented and first-generation college students in genuine research experiences.
Silencing the expression of a gene by inducing an RNA interference (RNAi) response is a common approach for performing studies of gene function in invertebrates. However, delivering enough double-stranded RNA to achieve sufficient gene knockdown through injection or feeding is expensive and ineffective in many insects, including bees. In this project, a FUnctional Genomics Using Engineered Symbionts (FUGUES) methodology will be developed and applied to honey bees (Apis mellifera) and bumblebees (Bombus spp.). In FUGUES, microbial symbionts are engineered to continuously produce and deliver double-stranded RNA to induce a targeted RNAi response in their host. Newly emerged bees colonized with an engineered bacterial symbiont exhibit reduced expression of a target gene throughout the bee body, enabling one to ascertain the function of a bee gene and its role in determining specific phenotypes. There are key advantages of using FUGUES to study gene function over current techniques that generate transgenic animals: it can be accomplished more quickly, it can be conducted in high-throughput when coupled with insect colonization via feeding, and it can be applied to species, such as bees, with mating systems and collective behaviors that complicate using genome editing techniques. Improving and disseminating the FUGUES tools created in this work will broadly enable studies of genes underlying bee behavior, development, and physiology. These tools will likely also be useful for studying many other insect species and other organisms that harbor symbiotic bacteria.
Selected publications
- Phillips LE, Sotelo KL, Moran NA. 2024. Characterization of gut symbionts from wild-caught Drosophila and other Diptera: description of Utexia brackfieldae gen. nov., sp. nov., Orbus sturtevantii sp. nov., Orbus wheelerorum sp. nov, and Orbus mooreae sp. nov. Int J Syst Evol Microbiol. 74(9):006516. doi: 10.1099/ijsem.0.006516.